DNA STRUCTURE
Concepts:
|
To
consider the structure of DNA, we have to look at the subunit molecules used
to make DNA. An
important concept at this point is to realize that DNA, RNA, and proteins are
all made from smaller molecules that we can call subunits or building blocks. |
|
|
Macromolecule |
Subunit or building block |
|
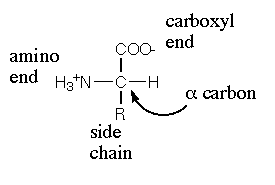
Proteins |
Amino Acids
|
|
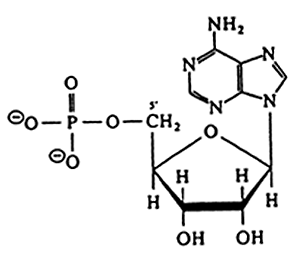
RNA |
Ribonucleotides
http://www.gravitywaves.com/chemistry/CHE%2520450/15_NucleotidesLipids.htm |
|
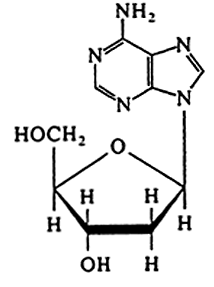
DNA |
Deoxyribonuleoside (no phosphate)
http://www.gravitywaves.com/chemistry/CHE%2520450/15_NucleotidesLipids.htm |
|
·
DNA is composed of 4 different deoxyribonucleotides:
Adenine Guanine, Cytosine, and Thymidine. We usually use the letters A, G, C, and T
to denote these subunits. ·
These subunits are attached together based upon a
base-pairing rule: o
Adenine pairs with Thymidine o
Guanine pairs with Cytosine ·
The synthesis of DNA requires a template DNA
molecule which is used to direct the insertion of the correct nucleotide into
the new molecule. This synthesis is
under the direction of DNA polymerase enzymes. ·
The typical structure of DNA is the Watson-Crick model where there are two strands of DNA
paired together by hydrogen bonds between the nucleotide bases. |
|
|
|
|
http://mrsec.wisc.edu/edetc/DNA/images/DNA3-5.jpg
Activity:
DNA structure will be
demonstrated by showing students how to read a sequencing gel. The sequencing gel is produced when DNA
strands are separated in an electric field.
DNA is negative charged (the phosphates are negative charged thus the
entire DNA molecule acts like a negative-charged rod). In an electric field, DNA molecules will
migrate toward the positive charged area.
If the DNA is placed in a semi-solid slab of agarose
(made from seaweed), the DNA molecules will be separated based upon their
sizes; the smaller the size, the further toward the positive pole the DNA
will migrate. This is called electrophoresis. DNA Electrophoresis
Electrophoresis is the most
common technique used in laboratories that work with DNA. Usually, the DNA samples are loaded in the
gel along side a known standard that has DNA fragments of known sizes. After the electrophoresis, the size of
unknown DNA molecules can be estimated by comparing to the standard. This comparison can be done
by measuring the distance of migration for each standard DNA molecule and
graphing the distance migrated vs the size (the
number of bases in the DNA, denoted as base pairs (bp)). This graph of the standard can then be used
to estimate the size of each unknown DNA molecule by comparing the distance
migrated to the standard graph. Material to perform DNA
electrophoresis can be ordered from various vendors. As an example; EDVOTEK has both agarose
gel electrophoresis kits (M36 HexaGel Horizontal
Electrophoresis Apparatus for $295). In addition, DNA samples and standards are
also available (samples for 6 gels with
methlyene blue stain for $61) Determining the Sequence of a DNA Molecule
The sequence of a DNA
molecule (the identity of each base found in the DNA) can be determined by
combining electrophoresis with a method called Dideoxynucleotide
Synthesis. In this method, the DNA
molecule to be sequenced is divided into four tubes, each tube contains an
enzyme called DNA polymerase (this enzyme is used to synthesize a copy of the
DNA molecule), the four deoxynucleotides (A, T, G,
and C), and a short primer that will bind to the DNA molecule to support the
synthesis. Also, each tube contains a
different dideoxynucleotide. This nucleotide lacks
a hydroxyl group on carbon number 3 of the ribose, thus if this dideoxynucleotide is incorporated into the synthesizing
DNA strand, no more nucleotides can be added.
The incorporation of this dideoxynucleotide
results in termination of the DNA synthesis reaction. An animation of this
reaction can be viewed at: Genetic Science
Learning Center (requires Macromedia Flash Player) Virtual
Lab: Agarose Electrophoresis Electrophoresis
- Topic Introduction After the reactions are
finished, each tube is loaded onto a polyacryamide
gel (like agarose, this is a semi-solid matrix that
is usually poured into a small diameter long tube and allowed to solidify.
With polyacrylamide, smaller DNA strands can be
separated in such a way that each band differs only from the next band by 1
nucleotide base. By reading the bands
starting from the bottom, the identity of the base can be determined. Activity I: 7th, 8th,
and 9th grade
We are using DNA sequencing
gels provided by EVOTEK. This activity requires the student
to take a sequencing gel and read the sequence. Notice that there are 4 lanes labeled A, T,
G and C. Starting from the bottom, find the band close to the bottom and
determine its lane. If it is lane A,
then A is the first base of the sequence.
Continue across the gel, going up the gel, finding the next largest
band. Determining its lane, write
down that letter as the next base.
Notice, as you get further up the gel, the bands gets closer together
so at one point it maybe difficult to distinguish between the bands. When you reach this point, you have read
all the sequence that can be read with accuracy. How many bases were you able to read?______________ Where any of your bases difficult to determine?________ What you have written is
the nucleotide sequence of one strand of DNA.
Using the base-pairing rules, can you write the sequence of the other
strand (remember DNA is double stranded)? |
Activity II: 9th-12th grade
Once a sequence has been determined
for a DNA molecule, it is usually desirable to determine what protein or RNA
molecule the DNA fragment codes for.
This is accomplished by making use of Internet-based databases. These databases contain sequences of genes
that have been previously identified.
The most useful database is maintained by the National Institutes of
Health (NIH) in Within the NCBI site are
many different databases; the main one is called Genbank. There are also databases of genomic
sequences such as human, drosophila, mouse, and many different bacteria. If you have a DNA sequence
you need to identify, you can search Genbank by
using the BLAST program. At the BLAST site, you will input your
sequence that you read from the gel and allow the BLAST program to search the
database to see if that sequence has been identified. The result obtained from the BLAST analysis
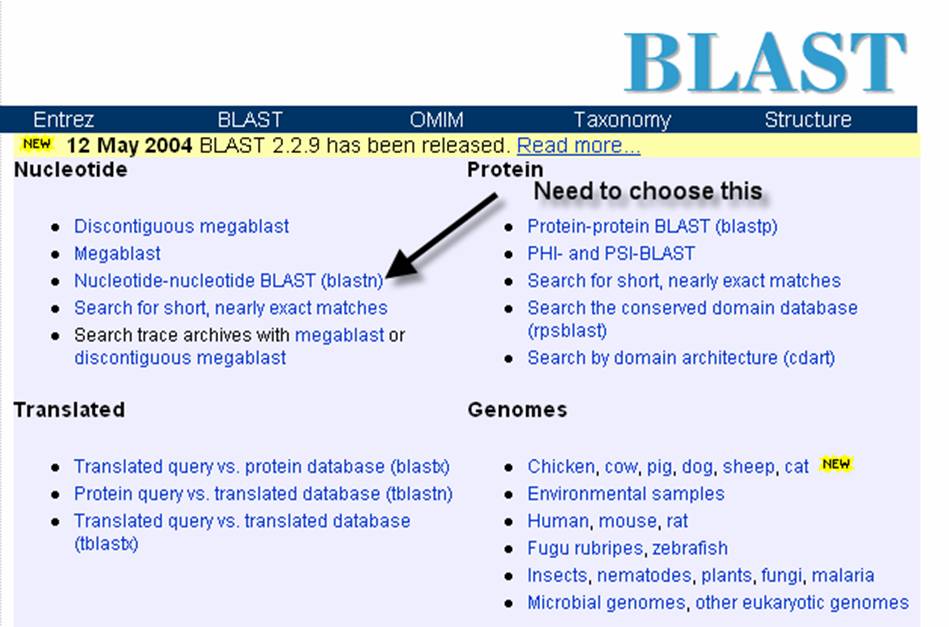
might give you a possible identity for your sequence. Once at the main BLAST
page, you will need to choose BLASTN program:
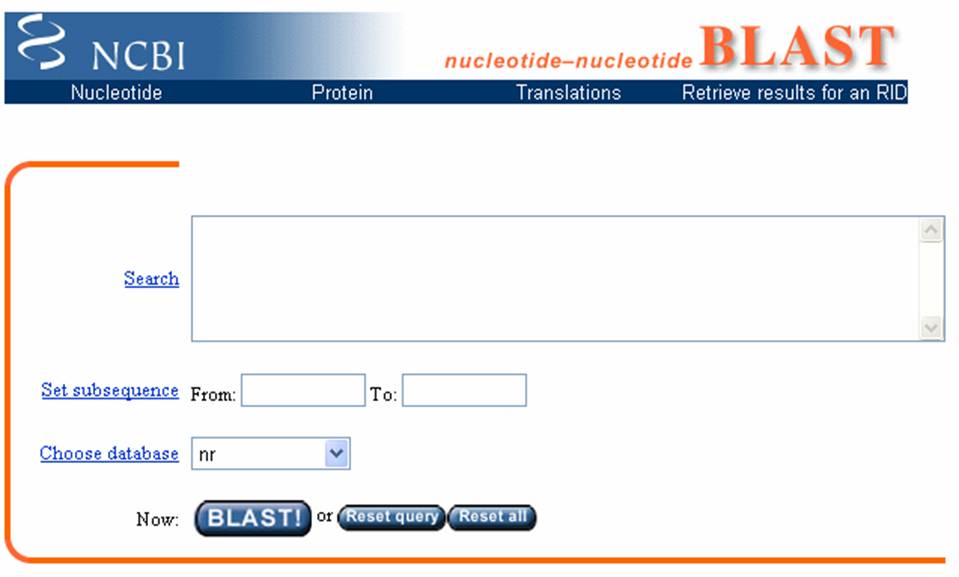
Observe the image below, this
image shows the entry portal to the BLAST site. In the marked box is where you will type
your unknown sequence. After entering
the sequence, push the BLAST button.
Another web page will open that has you task identifier and then you
will push the FORMAT button. Usually a
third web page opens that tells you the number of seconds you have to wait to
use the BLAST server (you actually are waiting in line to use the server).
When your sequence gets to the server, your results will be returned in
another web page.
Below is a
image of a BLAST result page. Notice
the list of sequences with links in blue.
These links will take you to other parts of the database to get more
information. The identity of your unknown
sequence is usually that sequence that is toward the top of the list. The interpretation of this analysis would
allow you to say “my sequence is similar to ____________________ that is
found in _____________ organism.
Determining the significance
of your analysis requires you to note the E value column. If you happen to
get a perfect match, then the E value would be 0.0 which means a perfect
match. Usually, the E value is
expressed as e-xxx. In fact, if the E value is less that e-0.02
the match between your unknown sequence and the database sequence is
significant. What is the possible identity for your sequence you read
off the gel? _______________________________________________________ Is this result considered significant? _____________ What
is the E value? _________________________ |