RESEARCH AREA
I) Synthetic/Medicinal Chemistry
- Structure-based design, synthesis and evaluation of inhibitory activity on β-Lactamase and inhibitors.
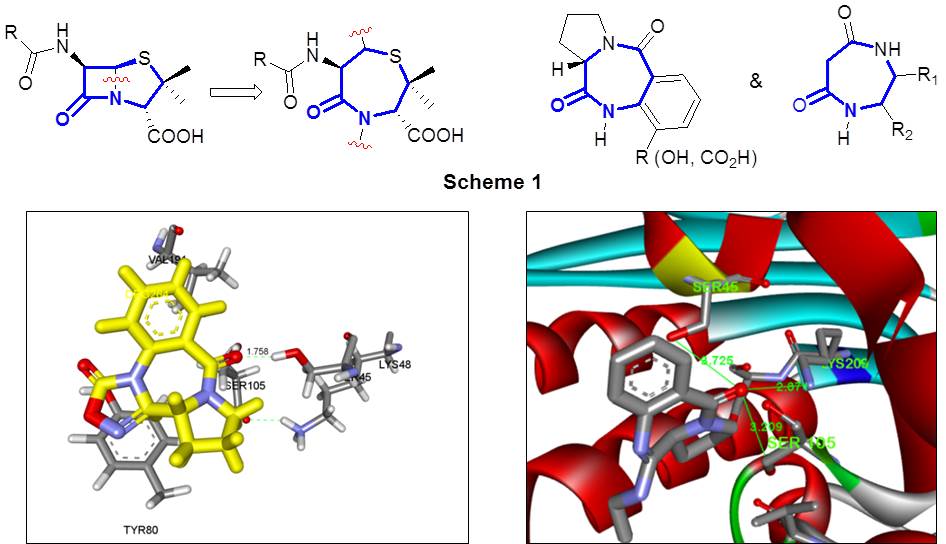
- Synthesis and In-Vitro Cell Viability/Cytotoxicity Studies of Novel pyrrolo[2,1-c][1,4] benzodiazepines (PBDs).
Currently, a continuing effort is being directed in our research lab towards the exploration and lead optimization of the PBD class of compounds and its closely related analogs. Further synthesis and structure-activity relationship (SAR) studies of the parent natural product and its tetracyclic analogs will lead to the discovery of established drug candidates. The synthesized PBD derivatives are being submitted to NCI Development Therapeutics Program (DTP)-Drug Synthesis and Chemistry Branch for NCI-60 cell lines in vitro cytotoxicity screening.
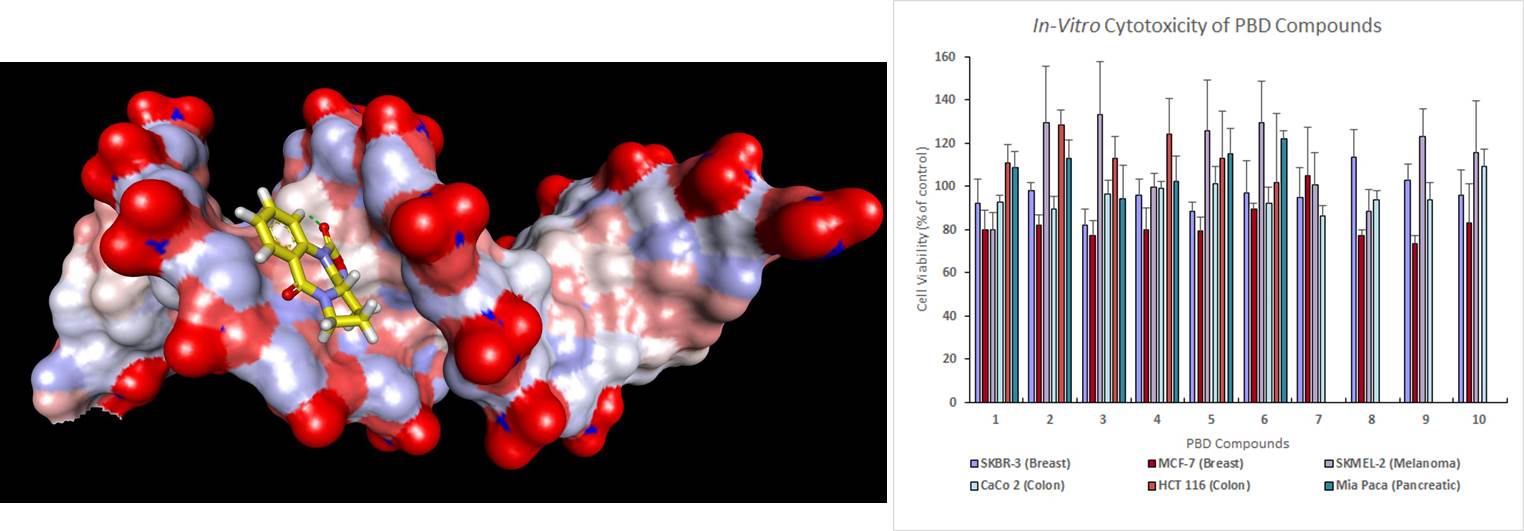
II) Natural Products Chemistry
- Isolation, purification, and structure determination of natural products from medicinal plant , marine species.
Total synthesis, semi-synthesis and structure-activity relationship studies of medicinally interest natural products and further lead-optimization of their established biological activity.
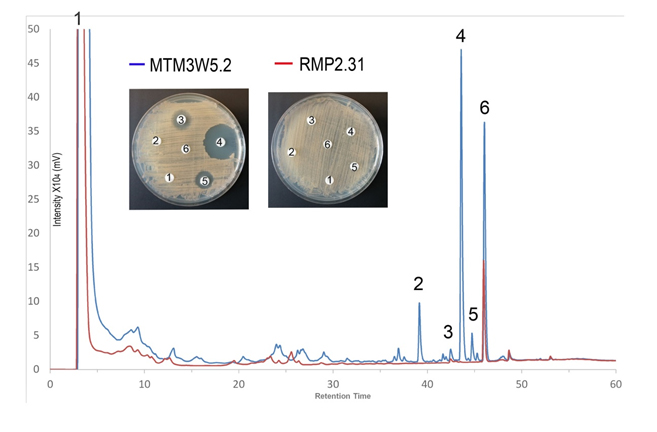
- Isolation and characterization of antibiotics produced by Rhodococcus soil bacterium.
The aim of this study is to isolate and characterize these new antibiotics produced by Rhodococcus strain MTM3W5.2. This will be done by extraction, purification and evaluation of secondary metabolites produced by the organism using HPLC, LCMS and MIC bioassays.
III) Modeling/Computational Chemistry
Computer-assisted design of efficient inhibitors using online programs, ZINC database virtual screening of more than 13 million commercially available compounds( http://zinc.docking.org ), QSAR/QSPR studies, as well as docking of individual molecules of interest. The crystal structures of β-lactam antibiotic-enzyme complexes are accessible from an online protein data bank.